
How does restriction endonuclease function?
Answer
591.6k+ views
Hint: Recombinant DNA technology (genetic engineering) exploits recombinant DNA as an important tool. A recombinant DNA molecule is made up of a vector DNA into which the desired DNA fragment has been inserted by using specific enzymes for cutting the DNA into suitable fragments and for ligation of the appropriate fragments.
Complete answer:
Endonucleases are the enzymes that produce internal cuts in DNA molecules. Many endonucleases cleave DNA molecules at random sites, but, there is a class of endonucleases that cleaves the DNA only within or near the sites which have specific base sequences. Such endonucleases are called Restriction Endonucleases and the sites recognized by them are called Recognition sites or recognition sequences. Restriction Endonucleases are commonly called as “molecular scissors” and are essential for Recombinant DNA technology.
Each Restriction Endonuclease functions by examining the length of a DNA sequence. When it identifies its specific recognition sequence, it binds to the DNA and cuts the DNA molecule by catalyzing the hydrolysis of the phosphodiester bond between adjacent nucleotides. To cut DNA, all restriction enzymes make two cuts, one in each of the two strands of the DNA double helix at specific points in their sugar-phosphate backbones.
A striking characteristic of the recognition sites is that they almost always possess two-fold-rotational symmetry. It means the recognized sequence is an inverted repeat or a palindrome and the cleavage sites are positioned symmetrically. The palindrome in DNA is a sequence of nucleotide base pairs that reads the same on the two strands when the orientation of reading is kept the same. The recognition sequences of a restriction enzyme EcoR1 is shown below to understand the palindromic nucleotide sequence –
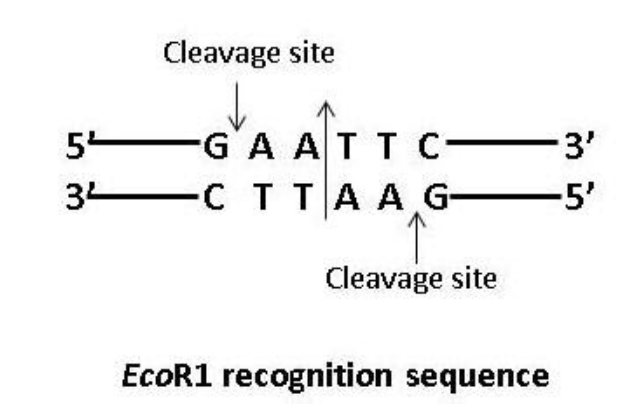
Figure 1. A palindrome sequence (The long arrow in the centre shows the axis of symmetry).
Restriction enzymes are used in genetic engineering for the formation of recombinant DNA molecules. The steps for the formation of recombinant DNA using restriction enzymes is shown in Figure 2. This includes the cleavage of vector DNA and desired DNA by the same restriction enzyme resulting in the same kind of sticky-ends. The sticky ends of vector DNA and the desired DNA fragment are joined together by an enzyme called DNA ligase.

Figure 2: Steps in the formation of Recombinant DNA
Note: There are three different types of Restriction Endonucleases –
(i) Type I, (ii) Type II and, (iii) Type III.
- Type I restriction endonucleases cleave DNA at random sites about more than 1,000 base pairs from the recognition sequence.
- Type III restriction endonucleases cleave the DNA about 25 base pairs from the recognition sequence. Both types, Type I and Type II require energy in the form of ATP for their activity.
- Type II restriction endonucleases are simple and do not require ATP, and cleave the DNA within the recognition sequence itself. Thus, only Type II Restriction Endonucleases are used for Recombinant DNA Technology.
Complete answer:
Endonucleases are the enzymes that produce internal cuts in DNA molecules. Many endonucleases cleave DNA molecules at random sites, but, there is a class of endonucleases that cleaves the DNA only within or near the sites which have specific base sequences. Such endonucleases are called Restriction Endonucleases and the sites recognized by them are called Recognition sites or recognition sequences. Restriction Endonucleases are commonly called as “molecular scissors” and are essential for Recombinant DNA technology.
Each Restriction Endonuclease functions by examining the length of a DNA sequence. When it identifies its specific recognition sequence, it binds to the DNA and cuts the DNA molecule by catalyzing the hydrolysis of the phosphodiester bond between adjacent nucleotides. To cut DNA, all restriction enzymes make two cuts, one in each of the two strands of the DNA double helix at specific points in their sugar-phosphate backbones.
A striking characteristic of the recognition sites is that they almost always possess two-fold-rotational symmetry. It means the recognized sequence is an inverted repeat or a palindrome and the cleavage sites are positioned symmetrically. The palindrome in DNA is a sequence of nucleotide base pairs that reads the same on the two strands when the orientation of reading is kept the same. The recognition sequences of a restriction enzyme EcoR1 is shown below to understand the palindromic nucleotide sequence –

Figure 1. A palindrome sequence (The long arrow in the centre shows the axis of symmetry).
Restriction enzymes are used in genetic engineering for the formation of recombinant DNA molecules. The steps for the formation of recombinant DNA using restriction enzymes is shown in Figure 2. This includes the cleavage of vector DNA and desired DNA by the same restriction enzyme resulting in the same kind of sticky-ends. The sticky ends of vector DNA and the desired DNA fragment are joined together by an enzyme called DNA ligase.

Figure 2: Steps in the formation of Recombinant DNA
Note: There are three different types of Restriction Endonucleases –
(i) Type I, (ii) Type II and, (iii) Type III.
- Type I restriction endonucleases cleave DNA at random sites about more than 1,000 base pairs from the recognition sequence.
- Type III restriction endonucleases cleave the DNA about 25 base pairs from the recognition sequence. Both types, Type I and Type II require energy in the form of ATP for their activity.
- Type II restriction endonucleases are simple and do not require ATP, and cleave the DNA within the recognition sequence itself. Thus, only Type II Restriction Endonucleases are used for Recombinant DNA Technology.
Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE

Draw a neat and well labeled diagram of TS of ovary class 12 biology CBSE




