What Is the Timeline and Importance of the Human Genome Project?
The Human Genome Project is considered one of the greatest scientific achievements in the field of biology. By sequencing the entire human genome, scientists unlocked the genetic code of our species, revealing essential details about genes, heredity, and disease. In this article, we’ll explore the journey, scientific foundations, process, and impacts of the Human Genome Project in a simple, student-friendly way.
What is the Human Genome Project?
The Human Genome Project (HGP) was a massive international research effort launched to map and sequence all the genes present in human DNA. The aim was to produce a comprehensive reference of the entire human genome, helping researchers understand gene function, inheritance, and the genetic basis of health and disease. This breakthrough created a foundation for innovations in genetics, medicine, agriculture, and many areas of science. The knowledge gained has benefited topics such as inherited and acquired traits, biotechnology, and disease diagnosis.
Science Behind the Human Genome Project
Understanding the Human Genome Project requires knowledge of classical, molecular, and human genetics. Classical genetics began with Gregor Mendel's experiments, which revealed basic inheritance laws. Later, scientists discovered that DNA and RNA are the carriers of genetic instructions. Each DNA molecule contains genes made up of sequences of nucleotides (adenine, thymine, guanine, and cytosine). These sequences serve as blueprints for all bodily proteins and processes.
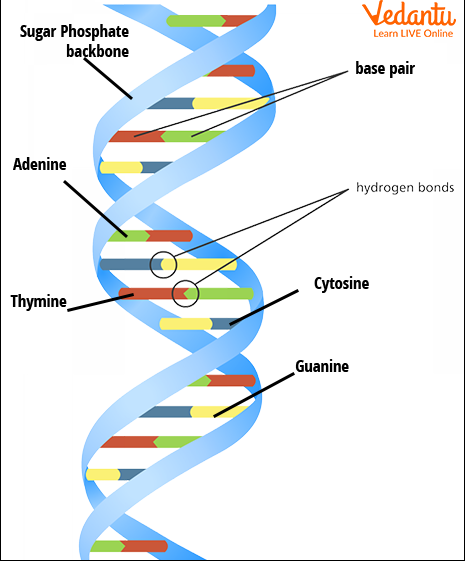
With discoveries such as Mendel’s laws, DNA structure, and gene regulation, researchers recognized the value in mapping the human genome. The Human Genome Project combined traditional genetics with molecular biology, computers, and advanced laboratory techniques to sequence the genome efficiently.
Timeline of the Human Genome Project
The Human Genome Project was a global collaboration involving scientists from the United States, United Kingdom, Japan, China, France, and other countries. It started in 1990 under the guidance of Francis Collins, with support from organizations like the National Institutes of Health and the U.S. Department of Energy. Over time, private companies also joined the race to sequence the genome, accelerating the pace of discovery and leading to technological advancements.
Key Milestones in the HGP
- 1990: Project officially begins, with a projected length of 15 years.
- 1998: Private company Celera Genomics enters and boosts competition.
- June 2000: First draft of the human genome sequence is completed.
- 2003: Human Genome Project is declared complete, two years ahead of schedule, coinciding with the 50th anniversary of DNA structure discovery.
Technological advances, such as automated sequencing and computational analysis, allowed researchers to read billions of DNA bases quickly and accurately.
Steps Used in the Human Genome Project
The process of sequencing the human genome followed several systematic steps. Specialized instruments and software handled massive amounts of DNA data. Here’s how the project worked:
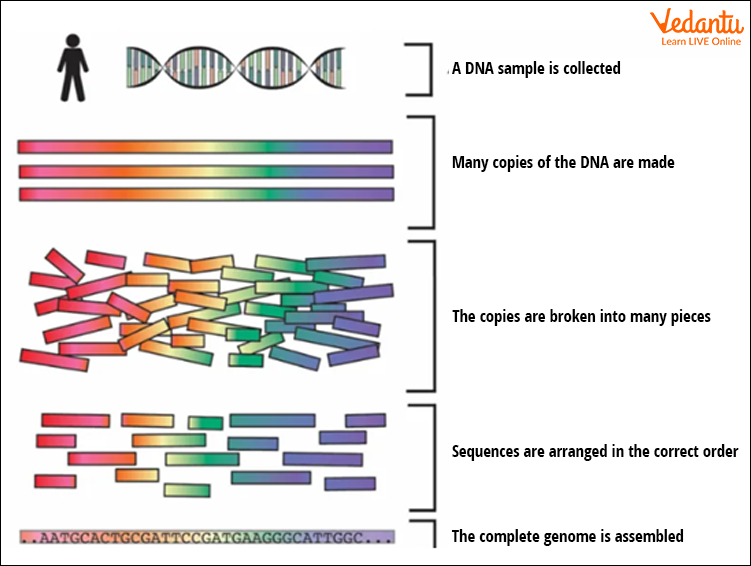
- DNA Extraction: Collecting DNA samples from volunteers.
- Fragmentation: Breaking DNA into manageable pieces for sequencing.
- Sequencing: Determining the order of bases (A, T, G, C) in each fragment.
- Assembly: Using computers to piece together overlapping sequences into a complete genome.
- Annotation: Identifying genes, coding regions, and non-coding regions.
- Analysis and Sharing: Making the data publicly available for further research and medical use.
This organized approach ensured every step, from extraction to sharing, was accurate, reliable, and transparent, setting standards followed in modern genetic research.
Outcomes and Significance of the Human Genome Project
The Human Genome Project revolutionized our understanding of the genetic basis of human biology. It showed that humans have around 20,000 to 25,000 genes. Much of the genome consists of non-coding DNA, with complex roles in gene regulation. The outcomes go far beyond science labs and have impacted many fields, including:
- Medicine: Improved diagnosis and treatment of genetic disorders.
- Pharmacogenomics: Tailoring medicines to individual genetic profiles.
- Agriculture: Enhanced breeding programs and genetically modified crops.
- Environmental studies: Understanding the effects of genetic diversity on adaptation (see climate effects).
- Evolution and inheritance: Better knowledge of how traits pass from one generation to the next (difference between acquired and inherited traits).
The HGP has also inspired new research in fields like genetic engineering, endocrinology, and nutrition. For example, deeper insights into genes related to hormones and metabolic processes help guide studies in endocrinology, food science, and human physiology.

Future Directions and Real-World Applications
The completion of the Human Genome Project opened doors to many new possibilities. Personalized medicine, gene therapy, early detection of inherited diseases, and understanding the genetics of diseases like cancer are all growing fields. The techniques pioneered during HGP are used in research about biomolecules, life science, and disease control. It also enables us to study adaptations, environmental challenges, and human evolution in more detail.
By following HGP’s legacy, Vedantu aims to deliver clear, accessible explanations for biology topics—making scientific advances understandable for every student.
Page Summary: The Human Genome Project was a landmark scientific effort that mapped the entire human genetic code. It revolutionized medicine, genetic research, and our understanding of hereditary diseases. Its outcomes influence health, evolution, agriculture, and environmental studies, with everyday relevance and ongoing innovations that continue to benefit both society and the scientific community.


FAQs on Human Genome Project Explained for Students
1. What is the Human Genome Project?
The Human Genome Project (HGP) is an international scientific research effort that aimed to map and sequence the entire DNA of humans. Its main objectives were to:
- Identify all the genes in human DNA
- Determine the sequences of the three billion base pairs
- Store this information in databases
- Improve data analysis tools
- Address ethical, legal, and social issues
2. What were the main goals of the Human Genome Project?
The main goals of the Human Genome Project (HGP) were to decode all human DNA and make this information available for scientific and medical research. Key objectives included:
- Mapping the human genome by identifying and locating all genes
- Sequencing the complete DNA base pairs
- Storing and analyzing genomic data
- Addressing ethical, legal, and social implications
3. Who were the key contributors to the Human Genome Project?
The Human Genome Project was a global collaboration led by several organizations and scientists. Major contributors included:
- National Institutes of Health (NIH), USA
- Department of Energy (DOE), USA
- Wellcome Trust, UK
- Celera Genomics (private company)
- International research teams from Japan, France, Germany, China, and others
4. What were the achievements of the Human Genome Project?
The Human Genome Project achieved several groundbreaking milestones in the field of genetics and biotechnology. Major achievements include:
- Mapping the locations of all human genes
- Sequencing approximately 99% of the human genome with high accuracy
- Creating public databases of genetic information
- Developing DNA analysis tools and techniques
- Promoting international scientific collaboration
5. Why is the Human Genome Project important?
The Human Genome Project is important because it expanded our understanding of human genetics and revolutionized medicine. Its significance can be summarized as:
- Enabling gene-based diagnosis and treatment
- Identifying genetic risk for diseases
- Facilitating research into gene therapy and personalized medicine
- Improving forensic science and anthropology
6. When was the Human Genome Project completed?
The Human Genome Project was officially completed in April 2003 after thirteen years of work. The draft version was published in 2001, with final details and improvements continuing until 2003, marking a historic achievement in the life sciences.
7. What are the ethical, legal, and social issues related to the Human Genome Project?
Ethical, legal, and social issues (ELSI) are a significant aspect of the Human Genome Project. Key concerns include:
- Privacy and confidentiality of genetic information
- Potential for genetic discrimination by employers or insurers
- Informed consent for genetic testing
- Impact on family relationships and society
- Ownership and patenting of genes
8. How did the Human Genome Project help in identifying diseases?
The Human Genome Project aided in identifying diseases by mapping genes and discovering genetic variations associated with health conditions. It helped by:
- Creating reference genome sequences for comparison
- Enabling identification of mutations linked to diseases
- Supporting early diagnosis and risk assessment
- Guiding the development of targeted therapies and gene-based treatments
9. What are some applications of the Human Genome Project in medicine?
Applications of the Human Genome Project in medicine are diverse and transformative. They include:
- Personalized medicine tailored to genetic profiles
- Gene therapy for inherited disorders
- Pharmacogenomics (predicting drug responses)
- Early detection and prevention of diseases
- Genetic counseling and enhanced forensic techniques
10. What is meant by genome mapping and sequencing?
Genome mapping and sequencing are two vital steps in genomics.
- Genome mapping refers to locating genes and markers on chromosomes
- Genome sequencing involves determining the exact order of DNA bases
11. How has the Human Genome Project impacted biological research?
The Human Genome Project has transformed biological research by providing comprehensive genetic data and advanced tools. Its impact includes:
- Accelerating discoveries in genomics and molecular biology
- Enabling cross-species comparisons and evolutionary studies
- Informing research in agriculture and biotechnology
12. Describe the role of bioinformatics in the Human Genome Project.
Bioinformatics played a critical role in the Human Genome Project by managing and analyzing large-scale genetic data. Important contributions were:
- Developing databases for storing genome sequences
- Creating computational tools for gene analysis
- Automating data processing and visualization
- Facilitating sharing of genetic information globally










