What Are the Essential Steps in Recombinant DNA Technology?
Recombinant DNA Technology is a pivotal innovation in biotechnology that has revolutionised medicine, agriculture, and research. In this article, we will explore recombinant dna technology steps, delve deeper into the concept of gene cloning, and look at recombinant dna technology examples to illustrate its real-world applications. We will also learn about restriction enzymes and their critical role in the entire process. By the end of this page, you will have a comprehensive overview of the 7 steps in recombinant dna technology, along with interactive activities to reinforce your knowledge.
What is Recombinant DNA Technology?
Recombinant dna technology (often called genetic engineering) involves merging DNA fragments from different biological sources to create a novel genetic sequence. This new DNA is then introduced into a host organism’s genome, enabling the host to express the inserted genes. The result can be anything from synthesising human insulin in bacteria to creating crop varieties with enhanced nutritional content or disease resistance.
Key highlights:
Combines DNA from different species or organisms.
Often relies on vectors (like plasmids or bacteriophages) to deliver desired genes.
Uses a series of restriction enzymes and other molecular tools to modify and integrate DNA.
Widely applied in medicine, agriculture, diagnostics, and research.
Essential Tools in Recombinant DNA Technology
Restriction Enzymes
These molecular “scissors” recognise specific DNA sequences and cut the DNA at or near these sites.
Endonucleases cut within the DNA strand, whereas exonucleases remove nucleotides from the ends of the strand.
Commonly recognise palindromic sequences, generating either “sticky ends” or “blunt ends.”
Critical for preparing both the vector DNA and the gene of interest so they can be joined later.
Vectors
Carriers that transport the desired DNA into the host cell.
Plasmids (circular DNA in bacteria) and bacteriophages are popular choices due to their high copy number and ease of manipulation.
Contain an origin of replication (for DNA duplication), a selectable marker (often an antibiotic-resistance gene), and a cloning site (recognition site for restriction enzymes).
Polymerases and Ligases
DNA Polymerases: Extend the DNA strand during processes like PCR (Polymerase Chain Reaction).
DNA Ligase: The “glue” that seals the nicks in the sugar-phosphate DNA backbone, forming stable recombinant DNA.
Host Organism
The cell into which the recombinant DNA is introduced. Common hosts include bacteria (e.g. E. coli), yeast, or even plant and animal cells in more complex procedures.
Must be able to replicate the inserted DNA and allow for gene expression.
Additional Techniques
PCR (Polymerase Chain Reaction): Amplifies DNA fragments to produce millions of copies from a single template.
Transformation Methods: Techniques such as microinjection, biolistics (gene gun), heat shock with calcium chloride, or electroporation to introduce recombinant DNA into the host.
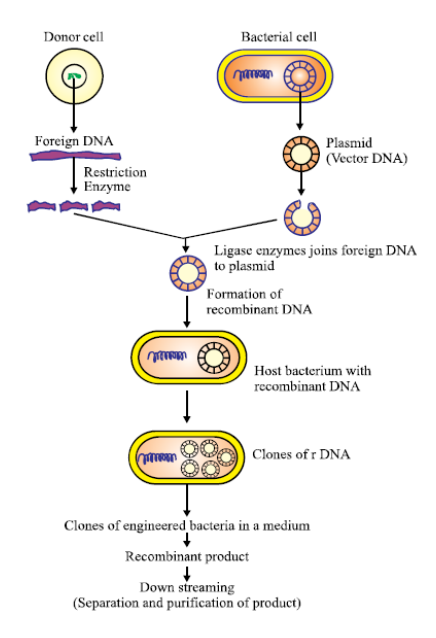
The 7 Steps in Recombinant DNA Technology
Although recombinant dna technology steps can vary slightly depending on the specific application, here are the 7 steps in recombinant dna technology typically followed:
Isolation of Genetic Material
Extract DNA from the donor organism in a pure form, removing proteins, lipids, and other macromolecules.
Cutting the DNA with Restriction Enzymes
Use appropriate restriction enzymes to cut both the donor DNA (gene of interest) and the vector at specific sites.
Ensures complementary sticky ends or blunt ends for ligation.
Amplification of Desired Gene
Employ PCR to exponentially increase the number of copies of the gene of interest, making downstream processes more efficient.
Ligation of DNA Fragments
Join the cut fragment of donor DNA with the vector using DNA ligase.
Creates a stable recombinant plasmid or vector that contains the desired gene.
Introduction of Recombinant DNA into the Host
Also known as transformation (in bacteria) or transfection (in mammalian cells).
Methods include heat shock, electroporation, or gene gun, depending on the host cell.
Screening/Selection of Transformants
Identify and select the host cells that have successfully taken up the recombinant DNA.
Often uses selectable markers (e.g., antibiotic resistance). Only transformants with the plasmid survive on antibiotic-containing media.
Expression and Maintenance in the Host
Under favourable conditions, the host cell replicates the recombinant plasmid and expresses the inserted gene.
The product (e.g., protein or hormone) is then harvested, and the recombinant gene can be passed on to daughter cells.
Recombinant DNA Technology Examples
Production of Synthetic Human Insulin: Bacteria engineered with the human insulin gene can produce insulin for diabetes management.
HIV Detection: Recombinant DNA-based diagnostics are used in rapid detection and monitoring of the virus.
Genetically Modified Crops: Bt-cotton and “Flavr Savr” tomatoes are recombinant dna technology examples that showcase improved pest resistance and shelf life.
Golden Rice: Engineered to synthesise beta-carotene, addressing Vitamin A deficiency in many parts of the world.
Gene Cloning – Creating Multiple Copies
Gene cloning is the process of producing numerous identical copies of a specific gene or DNA segment. In gene cloning:
A fragment of interest is inserted into a cloning vector.
The vector is introduced into a host like bacteria.
As bacteria multiply, they create millions of copies of the inserted gene.
This method is crucial for studying gene structure, function, and for producing substances like hormones, enzymes, and vaccines.
Applications of Gene Cloning
Medicine: Synthesis of hormones (e.g., insulin), antibiotics, and vitamins.
Agriculture: Development of stress-resistant and high-yield crops, often by integrating genes for pest or disease resistance.
Gene Therapy: Potential to fix defective genes by introducing functional copies into a patient’s cells.
Research: Understanding genetic pathways and disease mechanisms by studying cloned genes.
Also, read Gene Therapy
How Is This Different from DNA Cloning?
While both terms are closely related, “DNA cloning” often refers to the broader technique of replicating any fragment of DNA (possibly non-coding). Gene cloning specifically focuses on coding segments (functional genes). Essentially, gene cloning is a subset of DNA cloning where the target is a functional gene.
Ethical and Safety Considerations
One aspect that is often overlooked is the ethical and regulatory framework surrounding recombinant dna technology. There are guidelines to prevent the misuse of genetically modified organisms (GMOs) and to ensure that products entering the market are safe for both human use and the environment. Ongoing research also emphasises biosafety measures, transparent labelling of GMO foods, and public awareness.
Explore Genetic Disorders
Interactive Quiz on Recombinant DNA Technology
Test your understanding with this short quiz:
Which enzyme is primarily responsible for sealing the DNA backbone during ligation?
a) DNA Polymerase
b) DNA Ligase
c) Reverse Transcriptase
Identify the correct order of the 7 steps in recombinant dna technology from the options below:
a) Ligation → Isolation → Amplification → Cutting → Insertion → Screening → Expression
b) Isolation → Cutting → Amplification → Ligation → Insertion → Screening → Expression
c) Isolation → Amplification → Screening → Cutting → Insertion → Ligation → Expression
Which of the following is NOT a method of introducing recombinant DNA into a host cell?
a) Biolistics
b) Heat Shock
c) Transcription
Restriction enzymes typically recognise which type of sequences in DNA?
a) Single-stranded
b) Palindromic
c) Repetitive
Check Your Answers
b) DNA Ligase
b) Isolation → Cutting → Amplification → Ligation → Insertion → Screening → Expression
c) Transcription
b) Palindromic


FAQs on Recombinant DNA Technology: A Student’s Guide to Genetic Engineering
1. What is recombinant DNA technology in simple terms?
Recombinant DNA (rDNA) technology is a scientific method used to join together DNA fragments from two different sources. This creates a new, hybrid DNA molecule. This recombinant DNA is then introduced into a host organism, like a bacterium, to produce a specific protein or create a new trait.
2. What are the key steps involved in the process of creating recombinant DNA?
The process generally involves several core steps:
- Isolation of DNA: Extracting the desired gene from the source organism and the vector DNA (like a plasmid) from another.
- Cutting the DNA: Using the same restriction enzyme to cut both the desired gene and the vector DNA, creating compatible 'sticky ends'.
- Ligation: Joining the gene and the vector DNA together using an enzyme called DNA ligase. This forms the recombinant DNA molecule.
- Transformation: Introducing the recombinant DNA into a suitable host cell (e.g., bacteria).
- Selection and Screening: Identifying and isolating the host cells that have successfully taken up the recombinant DNA.
3. What are some common real-world examples of recombinant DNA technology?
This technology has many practical applications, especially in medicine and agriculture. For example, it is used to produce large quantities of human insulin for treating diabetes, create vaccines, and develop genetically modified crops (like Bt cotton) that are resistant to pests.
4. What is the role of a 'vector' in recombinant DNA technology?
A vector acts as a delivery vehicle. It is a DNA molecule, typically a plasmid or a virus, used to carry the foreign gene of interest into a host cell. The vector must be able to replicate inside the host cell to make many copies of the inserted gene.
5. Why are plasmids from bacteria often chosen as vectors for gene cloning?
Plasmids are ideal vectors for several reasons. They are small, circular DNA molecules that can replicate independently of the bacterial chromosome, ensuring many copies of the inserted gene are made. They are also easy to isolate and manipulate in the lab and often contain selectable markers (like antibiotic resistance genes) which help identify which bacteria have successfully taken up the plasmid.
6. How do restriction enzymes, the 'molecular scissors', know exactly where to cut a DNA strand?
Restriction enzymes are highly specific. Each enzyme recognises and binds to a unique, short DNA sequence called a recognition site. These sites are often palindromic, meaning the DNA sequence reads the same forwards and backwards on opposite strands. The enzyme only cuts the DNA at this specific site, ensuring a precise and predictable cut.
7. What is the difference between 'sticky ends' and 'blunt ends', and why is it important in this technology?
When a restriction enzyme cuts DNA, it can leave two types of ends. Sticky ends are short, single-stranded overhangs that can easily pair with a complementary sticky end. Blunt ends are straight cuts with no overhangs. Sticky ends are generally preferred in recombinant DNA technology because they make it much easier for the foreign gene to attach to the vector DNA through complementary base pairing, increasing the efficiency of the ligation process.
8. What is the purpose of using PCR (Polymerase Chain Reaction) in the recombinant DNA process?
PCR is used for gene amplification. If you only have a very small amount of the desired gene to start with, PCR can be used to make millions or billions of copies of that specific DNA segment. This ensures there is enough DNA material to successfully insert it into vectors and carry out the experiment.
9. How do scientists know if the recombinant DNA was successfully inserted into a host cell?
Scientists use a technique involving selectable markers. These are special genes included in the vector, often providing resistance to an antibiotic. After transformation, the host cells (e.g., bacteria) are grown on a medium containing that antibiotic. Only the cells that have successfully taken up the vector with the resistance gene will survive, making them easy to identify and select.
10. Besides medicine, in what other fields is recombinant DNA technology making a significant impact?
Beyond medicine, this technology is revolutionising other areas. In agriculture, it's used to create crops with higher yields, better nutritional value, and resistance to diseases. In industry, it's used to produce enzymes for detergents and food processing. It is also a critical tool in environmental science for processes like bioremediation, where bacteria are engineered to clean up pollutants.










