
GAATTC is the recognition site for which of the following restrictions endonuclease?
a. Hind III
b. EcoR I
c. Bam II
d. Hae III
Answer
595.2k+ views
Hint: The recognition sequence of nucleic acid where the enzyme cuts are G/AATTC, which has a complementary and palindromic sequence of CTTAA/G. Restriction endonuclease is also called the restriction enzyme. It cuts double helix DNA in a precise way.
Complete answer:
The species from which the restriction endonuclease enzyme, EcoR I, originated will state the end part of that enzyme.
Whereas the letter R is the representation of certain strains in this case it is RY13.
Figure – 2: structure of EcoRI
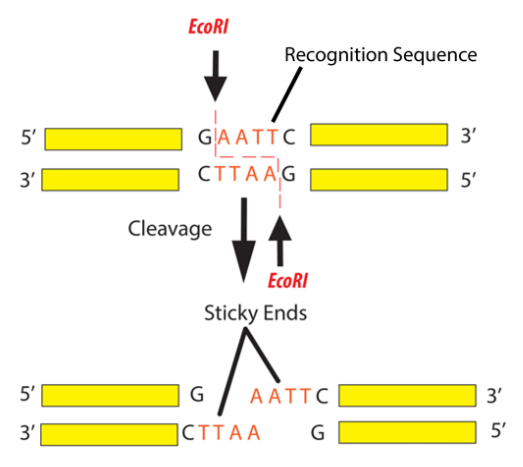
From the species E. coli, EcoR I is a restriction endonuclease enzyme that is isolated.
The number I which is the last part of its name, is the one which denotes that EcoR I was the first enzyme which is isolated from this straining.
The only restriction enzyme which cleaves the DNA double helices into fragments at precise positions is the EcoRI.
EcoRI is a restriction endonuclease enzyme which is also a part of the restriction-modification system.
The EcoR I is the endonuclease enzyme which is used as a restriction enzyme in molecular biology. EcoR I generate 4 nucleotide tacky split ends with 5' end ledges of AATT.
So, the correct option is 'EcoR I'.
Additional Information:
- A restriction enzyme (which is also called restriction endonuclease, or restrictive) can be termed as an enzyme that cleaves the complete DNA into fragments by the side of definite recognition locations within molecules known as restriction sites. The restriction enzymes are one class of the wider endonuclease group of enzymes.
- Hind III cleaves DNA at a particular site.
- The recurrent neural network that was introduced by Bart Kosko in 1988 is known as BAM (Bidirectional associative memory).
- The restriction enzyme that is of prokaryotic type of DNA and protects organisms from unknown, foreign DNA is called Hae III.
Note: In the above statements, the / in the classification designates the phosphodiester bond in the enzyme will disrupt the DNA particle. GAATTC is a palindromic sequence and can be defined as a sequence of the nucleic acid in a DNA or RNA molecule which is stranded double in which reading is in a firm direction.
Complete answer:
The species from which the restriction endonuclease enzyme, EcoR I, originated will state the end part of that enzyme.
Whereas the letter R is the representation of certain strains in this case it is RY13.
Figure – 2: structure of EcoRI

From the species E. coli, EcoR I is a restriction endonuclease enzyme that is isolated.
The number I which is the last part of its name, is the one which denotes that EcoR I was the first enzyme which is isolated from this straining.
The only restriction enzyme which cleaves the DNA double helices into fragments at precise positions is the EcoRI.
EcoRI is a restriction endonuclease enzyme which is also a part of the restriction-modification system.
The EcoR I is the endonuclease enzyme which is used as a restriction enzyme in molecular biology. EcoR I generate 4 nucleotide tacky split ends with 5' end ledges of AATT.
So, the correct option is 'EcoR I'.
Additional Information:
- A restriction enzyme (which is also called restriction endonuclease, or restrictive) can be termed as an enzyme that cleaves the complete DNA into fragments by the side of definite recognition locations within molecules known as restriction sites. The restriction enzymes are one class of the wider endonuclease group of enzymes.
- Hind III cleaves DNA at a particular site.
- The recurrent neural network that was introduced by Bart Kosko in 1988 is known as BAM (Bidirectional associative memory).
- The restriction enzyme that is of prokaryotic type of DNA and protects organisms from unknown, foreign DNA is called Hae III.
Note: In the above statements, the / in the classification designates the phosphodiester bond in the enzyme will disrupt the DNA particle. GAATTC is a palindromic sequence and can be defined as a sequence of the nucleic acid in a DNA or RNA molecule which is stranded double in which reading is in a firm direction.
Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

What is a transformer Explain the principle construction class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE




