
The normal function for restriction endonuclease is to
(a) Excise introns from hnRNA
(b) Polymerize nucleotides to form RNA
(c) Cleave and modify DNA
(d) Remove primer from Okazaki fragments
Answer
582.3k+ views
Hint: Endonucleases are enzymes that can break the phosphodiester bond present within a polynucleotide chain. a number of them haven't any reference to sequence when cutting DNA, but many others do so only at specific nucleotide sequences.
Complete step by step answer:
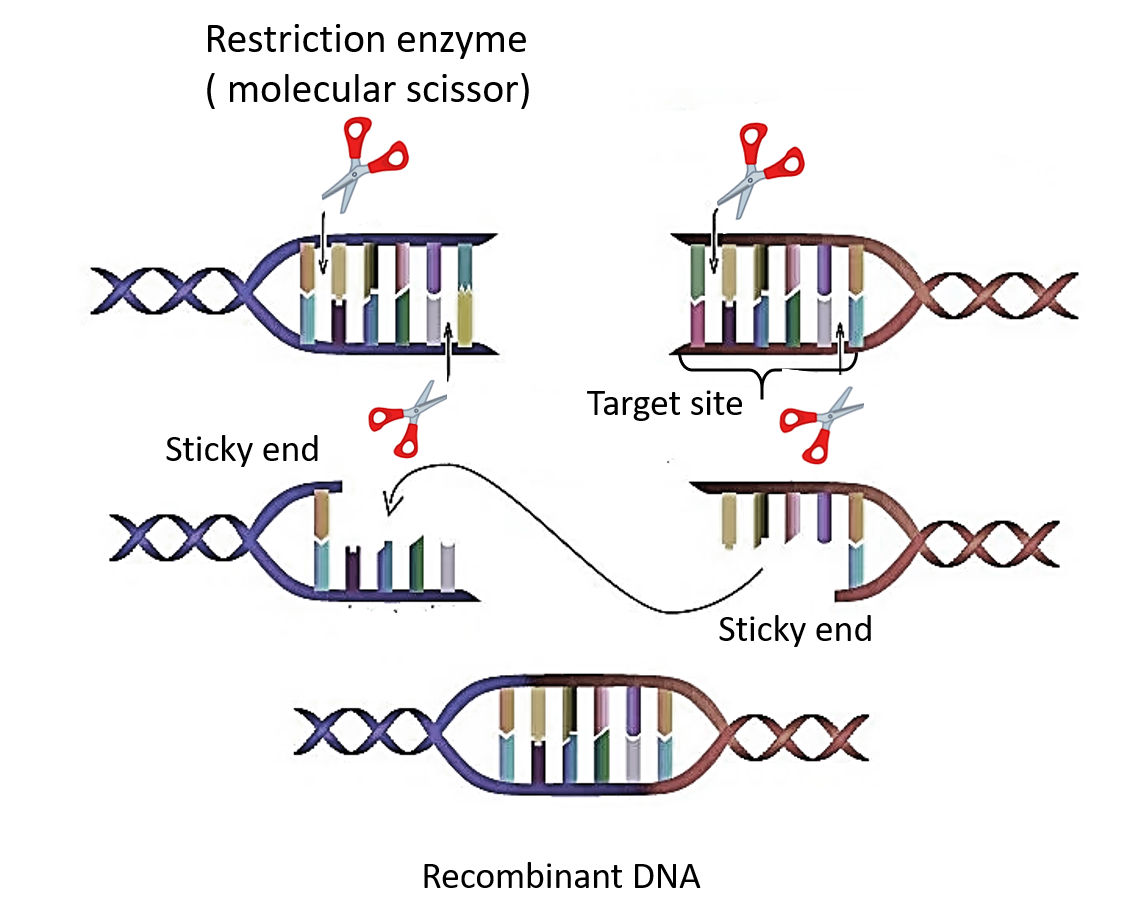
Restriction endonucleases identify the specific sequences within the DNA and digest the DNA into fragments by destroying the phosphodiester bonds. Hence, the restriction endonucleases are synthesised by bacteria as a part of their defence reaction which helps within the cleavage of the DNA segments which are identified as foreign. The restriction sites are very short sequences and may be present within the genome of the bacteria. Its own DNA is protected against destruction by adding methyl groups to it. Certain bases within the strand of DNA of the bacteria are modified for getting protected against the activity of the enzymes.
So, the correct answer is, ‘Cleave and modify DNA.’
Additional Information:
A restriction endonuclease is an enzyme that is isolated from the bacteria that can cut dsDNA into fragments. It does by recognizing specific nucleotide sequences as ‘recognition’ or ‘restriction site’. The term ‘restriction’ comes from the observation that these enzymes hamper or obstruct the entry of foreign DNA within the bacteria. EcoRII is isolated from gram-negative Escherichia coli.
The naming of restriction nucleases involves the abbreviations of the bacterial species from which the enzyme is isolated. E.g., Eco- for E. coli and Hin- for H. influenzae. Roman numerals also are used to indicate the sequence of discovery if more than one restriction endonuclease has been isolated from an equivalent bacterial strain.
Note:
- W. Arber was the very first scientist who postulated about ‘restriction enzymes.’ He noticed the fragmentation of bacteriophage’s DNA into smaller pieces when bacteriophage injects its DNA into the bacteria. In 1970, Hamilton-Smith and his coworkers first isolated a restriction enzyme from the bacterium Haemophilus Influenzae and named it HindII which recognizes a 6 pair dsDNA sequence.
- The action of restriction enzymes is either blunt or in staggered cuts. Blunt ends occur when both ends of cut DNA have a nucleotide. While in staggered cuts, sticky ends wherein the 2 cut ends of DNA don't possess any nucleotide.
Complete step by step answer:
Restriction endonucleases identify the specific sequences within the DNA and digest the DNA into fragments by destroying the phosphodiester bonds. Hence, the restriction endonucleases are synthesised by bacteria as a part of their defence reaction which helps within the cleavage of the DNA segments which are identified as foreign. The restriction sites are very short sequences and may be present within the genome of the bacteria. Its own DNA is protected against destruction by adding methyl groups to it. Certain bases within the strand of DNA of the bacteria are modified for getting protected against the activity of the enzymes.

So, the correct answer is, ‘Cleave and modify DNA.’
Additional Information:
A restriction endonuclease is an enzyme that is isolated from the bacteria that can cut dsDNA into fragments. It does by recognizing specific nucleotide sequences as ‘recognition’ or ‘restriction site’. The term ‘restriction’ comes from the observation that these enzymes hamper or obstruct the entry of foreign DNA within the bacteria. EcoRII is isolated from gram-negative Escherichia coli.
The naming of restriction nucleases involves the abbreviations of the bacterial species from which the enzyme is isolated. E.g., Eco- for E. coli and Hin- for H. influenzae. Roman numerals also are used to indicate the sequence of discovery if more than one restriction endonuclease has been isolated from an equivalent bacterial strain.
Note:
- W. Arber was the very first scientist who postulated about ‘restriction enzymes.’ He noticed the fragmentation of bacteriophage’s DNA into smaller pieces when bacteriophage injects its DNA into the bacteria. In 1970, Hamilton-Smith and his coworkers first isolated a restriction enzyme from the bacterium Haemophilus Influenzae and named it HindII which recognizes a 6 pair dsDNA sequence.
- The action of restriction enzymes is either blunt or in staggered cuts. Blunt ends occur when both ends of cut DNA have a nucleotide. While in staggered cuts, sticky ends wherein the 2 cut ends of DNA don't possess any nucleotide.
Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE

Draw a neat and well labeled diagram of TS of ovary class 12 biology CBSE




