
What is the 3'UTR site of mRNA?
Answer
521.4k+ views
Hint: Messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA. It carries the protein blueprint from a cell's DNA to its ribosomes to drive the protein synthesis. The structure of mRNA is in accordance with a gene encoded within DNA.
Complete answer:
mRNA is created during the process of transcription. In this process, the enzyme, RNA polymerase, converts the gene from DNA into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually includes introns, regions that will not go on to code of the final amino acid sequence. These are removed during RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes the mature mRNA which is read by the ribosome. With assistance of amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. Eukaryotic mRNA molecules often require considerable processing and transport, whereas prokaryotic mRNA molecules do not.
Coding regions constitute codons, which are decoded and translated into proteins by the ribosome. It begins with the start codon and ends with a stop codon. Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon. These regions are not translated and are termed the five prime untranslated regions (5' UTR) and three prime untranslated regions (3' UTR), respectively. The ability of a UTR to perform functions like mRNA stability, mRNA localization, and translational efficiency, depends on the sequence of the UTR and can differ between mRNAs. Because of the changes in RNA structure and protein translation, genetic variants in 3' UTR have a role to play in disease susceptibility. The stability of mRNAs may also be controlled by the 3' UTR and/or 5' UTR. Proteins that bind to either the 3' or 5' UTR may affect the process of translation by influencing the ribosome's ability to bind to mRNA. MicroRNAs bound to the 3' UTR may also affect translational efficiency or mRNA stability. The cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Therefore, the 3' UTR may contain sequences that allow the transcript to be localized to a particular region for translation.
In molecular genetics, the three prime untranslated region (3′ UTR) is the section of mRNA that immediately follows the end codon. By binding to specific sites within the 3′-UTR, microRNAs (miRNA) can cause a decrease in gene expression of various mRNAs. It happens so by either inhibiting the translation process or by directly causing the degradation of the transcript. The 3′ UTR also has silencer regions which bind to repressor proteins and inhibit the expression of mRNA. The 3′ UTR contains the sequence AAUAAA that directs the addition of several hundred adenine residues which are called the poly (A) tail, to the end of the mRNA transcript. The 3′ UTR can also contain sequences which attract proteins to link the mRNA with the cytoskeleton, transport it to or from the cell nucleus, or perform other types of localizations. The 3′-UTR also often contains AU-rich elements (AREs) to which are attached ARE-binding proteins (ARE-BP) which promote mRNA decay, affect mRNA stability, or activate translation. The length and secondary structure of the 3' UTR assists in the regulation of the translation process. Longer 3′- UTRs correspond to lower expression rates as they often contain more miRNA and protein binding sites which are involved in the inhibition of translation. Human 3' UTR are twice as long as the other mammals'.
3′-UTR mutations can not only affect alleles and physically linked genes but also unrelated genes since they are involved in the processing and nuclear export of mRNA. Mutations in AU-rich regions lead to diseases like tumorigenesis (cancer), hematopoietic malignancies and developmental delay/autism spectrum disorders. Elements in the 3′-UTR have also been associated with human acute myeloid leukemia, alpha-thalassemia, neuroblastoma, Keratinopathy, Aniridia, IPEX syndrome, and congenital heart defects.
Note:
mRNAs usually consist of several overlapping control elements. Therefore, it is often difficult to state and define the identity and function of each 3′-UTR element, let alone the regulatory factors that bind at these sites. Future research aided by the use of deep-sequencing based ribosome profiling, will reveal more regulatory subtleties in the 3' UTR region as well as any new control elements and AUBPs.
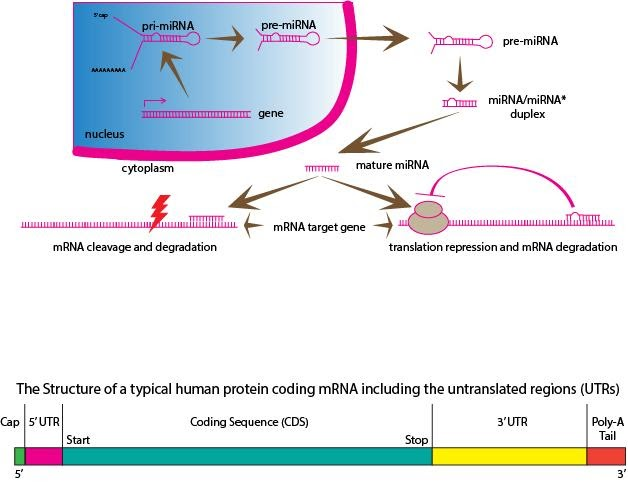
Figure 1: Coding mRNA
Complete answer:
mRNA is created during the process of transcription. In this process, the enzyme, RNA polymerase, converts the gene from DNA into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually includes introns, regions that will not go on to code of the final amino acid sequence. These are removed during RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes the mature mRNA which is read by the ribosome. With assistance of amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. Eukaryotic mRNA molecules often require considerable processing and transport, whereas prokaryotic mRNA molecules do not.
Coding regions constitute codons, which are decoded and translated into proteins by the ribosome. It begins with the start codon and ends with a stop codon. Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon. These regions are not translated and are termed the five prime untranslated regions (5' UTR) and three prime untranslated regions (3' UTR), respectively. The ability of a UTR to perform functions like mRNA stability, mRNA localization, and translational efficiency, depends on the sequence of the UTR and can differ between mRNAs. Because of the changes in RNA structure and protein translation, genetic variants in 3' UTR have a role to play in disease susceptibility. The stability of mRNAs may also be controlled by the 3' UTR and/or 5' UTR. Proteins that bind to either the 3' or 5' UTR may affect the process of translation by influencing the ribosome's ability to bind to mRNA. MicroRNAs bound to the 3' UTR may also affect translational efficiency or mRNA stability. The cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Therefore, the 3' UTR may contain sequences that allow the transcript to be localized to a particular region for translation.
In molecular genetics, the three prime untranslated region (3′ UTR) is the section of mRNA that immediately follows the end codon. By binding to specific sites within the 3′-UTR, microRNAs (miRNA) can cause a decrease in gene expression of various mRNAs. It happens so by either inhibiting the translation process or by directly causing the degradation of the transcript. The 3′ UTR also has silencer regions which bind to repressor proteins and inhibit the expression of mRNA. The 3′ UTR contains the sequence AAUAAA that directs the addition of several hundred adenine residues which are called the poly (A) tail, to the end of the mRNA transcript. The 3′ UTR can also contain sequences which attract proteins to link the mRNA with the cytoskeleton, transport it to or from the cell nucleus, or perform other types of localizations. The 3′-UTR also often contains AU-rich elements (AREs) to which are attached ARE-binding proteins (ARE-BP) which promote mRNA decay, affect mRNA stability, or activate translation. The length and secondary structure of the 3' UTR assists in the regulation of the translation process. Longer 3′- UTRs correspond to lower expression rates as they often contain more miRNA and protein binding sites which are involved in the inhibition of translation. Human 3' UTR are twice as long as the other mammals'.
3′-UTR mutations can not only affect alleles and physically linked genes but also unrelated genes since they are involved in the processing and nuclear export of mRNA. Mutations in AU-rich regions lead to diseases like tumorigenesis (cancer), hematopoietic malignancies and developmental delay/autism spectrum disorders. Elements in the 3′-UTR have also been associated with human acute myeloid leukemia, alpha-thalassemia, neuroblastoma, Keratinopathy, Aniridia, IPEX syndrome, and congenital heart defects.
Note:
mRNAs usually consist of several overlapping control elements. Therefore, it is often difficult to state and define the identity and function of each 3′-UTR element, let alone the regulatory factors that bind at these sites. Future research aided by the use of deep-sequencing based ribosome profiling, will reveal more regulatory subtleties in the 3' UTR region as well as any new control elements and AUBPs.

Figure 1: Coding mRNA
Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE

Draw a neat and well labeled diagram of TS of ovary class 12 biology CBSE




